AbstractDifferentiation.jl for AD-backend agnostic code
Differentiable programming (∂P), i.e., the ability to differentiate general computer program structures, has enabled the efficient combination of existing packages for scientific computation and machine learning1. The Julia2 language is well suited for ∂P, see also Chris’ article3 for a detailed examination. There is already a plethora of examples where ∂P has provided massive performance and accuracy advantages over black-box approaches to machine learning. This is because black-box machine learning approaches are flexible but require a large amount of data. Incorporating previously acquired knowledge about the structure of a problem reduces the amount of data and allows the learning task to be simplified4, for example, by focusing on learning only the parts of the model that are actually missing4 5. In the context of quantum control, we have demonstrated the power of this framework for closed6 and open quantum systems7.
∂P is (commonly) realized by automatic differentiation (AD), which is a family of techniques to efficiently and accurately differentiate numeric functions expressed as computer programs. Generally, besides forward- and reverse-mode AD, the two main branches of AD, a large variety of software implementations with different pros and cons exists. The goal is to make the best choice in every part of the program without requiring users to significantly customize their code. Having a common ground by ChainRules.jl empowers this idea of a Glue AD where backend developers just define ChainRules overloads. However, switching from one backend to another on the user side can still be tedious because the user has to look up the syntax of the new AD package.
Mohamed Tarek has started to implement a high level API for differentiation that unifies the APIs of all the AD packages in the Julia ecosystem. Ultimately, the API of our new package, AbstractDifferentiation.jl, aims at enabling AD users to write AD backend-agnostic code. This will greatly facilitate the switching between different AD packages. Once the interface is completed and all tests are added, it is also planned that DiffEqSensitivity.jl within the SciML software suite adopts AbstractDifferentiation.jl as a better way of handling AD choices. In this part of my GSoC project, I’ve started to fix remaining errors of the initial PR.
The interested reader is encouraged to look at Mohamed’s first PR for a complete list of functions provided by AbstractDifferentiation.jl (and some great discussions about the package). In the rest of this blog post, I will focus on a concrete example to illustrate the main idea.
Optimization of the Rosenbrock function
The Rosenbrock function is defined by
$$ g(x_1,x_2) = (a-x_1)^2 + b(x_2-x_1^2)^2. $$
The function $g$ has a global minimum at $(x_1^\star, x_2^\star)= (a, a^2)$ with $g(x_1^\star, x_2^\star)=0$. In the following, we fix $a = 1$ and $b = 100$. The global minimum is located inside a long, narrow, banana-shaped, flat valley, which makes the function a common test case for optimization algorithms.
Let us now implement the Gauss–Newton algorithm to find the global minimum. The Gauss–Newton algorithm iteratively finds the value of the $N$ variables ${\bf{x}}=(x_1,\dots, x_N)$ that minimize the sum of squares of $M$ residuals $(f_1,\dots, f_M)$
$$ S({\bf x}) = \frac{1}{2} \sum_{i=1}^M f_i({\bf x})^2. $$
Starting from an initial guess ${\bf x_0}$ for the minimum, the method runs through the iterations
$$ {\bf x}^{k+1} = {\bf x}^k - \alpha_k \left(J^T J \right)^{-1} J^T f({\bf x}^k), $$ where $J$ is the Jacobian matrix at ${\bf{x}}^k$ and $\alpha_k$ is the step length determined via a line search subroutine.
The following plot shows the Rosenbrock function in 3D as well as a 2D heatmap including the global minimum ${\bf x^\star}=(1,1)$ and our initial guess ${\bf x_0}=(0,-0.1)$.
using Pkg
path = @__DIR__
cd(path); Pkg.activate("."); Pkg.instantiate()
## AbstractDifferentiation is not released yet!!
using AbstractDifferentiation
using Test, LinearAlgebra
using FiniteDifferences, ForwardDiff, Zygote
using Enzyme, UnPack
using Plots, LaTeXStrings
# using Diffractor: ∂⃖¹ ## Diffractor needs >julia@1.6
## Rosenbrock function
# R: R^2 -> R: x -> (a-x₁)² + b(x₂-x₁²)²
g(x,p) = (p[1]-x[1])^2 + p[2]*(x[2]-x[1]^2)^2
# visualization
p = [1.0,100.0]
x₀ = [0.0,-0.1]
xopt = [1.0,1.0]
do_plot = true
if do_plot
x₁, x₂ = -2.0:0.01:2.0, -0.6:0.01:3.5
z = Surface((x₁,x₂)->g([x₁,x₂],p), x₁, x₂)
pl1 = surface(x₁,x₂,z, linealpha = 0.3, c=cgrad(:thermal, scale = :exp), colorbar=true,
labelfontsize=20,camera = (3,50),
xlabel = L"x_1", ylabel = L"x_2")
pl2 = heatmap(x₁,x₂,z, c=cgrad(:thermal, scale = :exp),
labelfontsize=20,
xlabel = L"x_1", ylabel = L"x_2")
scatter!(pl2, [(x₀[1],x₀[2])], label=L"x_0", legendfontsize=15, markershape = :circle, markersize = 10, markercolor = :green)
scatter!(pl2, [(xopt[1],xopt[2])],label=L"x^\star", legendfontsize=15, markershape = :star, markersize = 10, markercolor = :red)
pl = plot(pl1,pl2, layout=(2,1))
savefig(pl, "Rosenbrock.png")
end
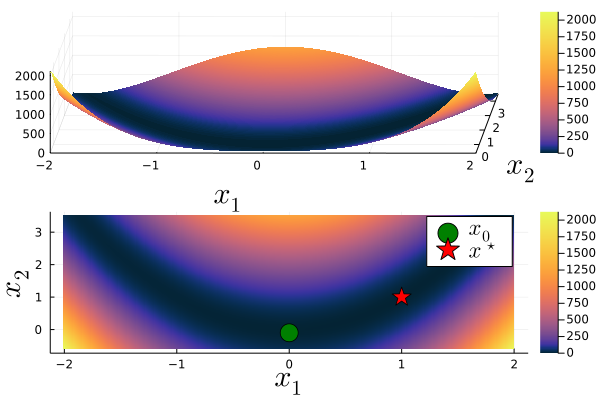
To apply the Gauss-Newton algorithm to the Rosenbrock function $g$, we first cast $g$ into an appropriate form fulfilling $S({\bf x})$, i.e., we use:
$$ f:\mathbb{R}^2\rightarrow\mathbb{R}^2: {\bf x} \mapsto \begin{pmatrix} f_1({\bf x}) \\ f_2({\bf x}) \\ \end{pmatrix} = \begin{pmatrix} \sqrt{2}(a-x_1) \\ \sqrt{2b}(x_2-x_1^2)\\ \end{pmatrix}, $$
instead of $g$. We can easily compute the Jacobian of $f$ manually
$$ J = \begin{pmatrix} -\sqrt{2} & 0 \\ -2x_1\sqrt{2b} & \sqrt{2b} \\ \end{pmatrix}. $$
We can then implement a (simple, non-optimized) version of the Gauss-Newton algorithm as follows.
# bring Rosenbrock function into the form "sum of squares of functions"
f1(x,p) = convert(eltype(x),sqrt(2))*(p[1]-x[1])
f2(x,p) = convert(eltype(x),sqrt(2*p[2]))*(x[2]-x[1]^2)
f(x,p) = [f1(x,p),f2(x,p)]
function f(res,x,p) # Enzyme works with inplace functions
res[1] = f1(x,p)
res[2] = f2(x,p)
return nothing
end
## manually pre-defined Jacobian
function Jacobian(x,p)
[-convert(eltype(x),sqrt(2)) 0
-2*x[1]*convert(eltype(x),sqrt(2*p[2])) convert(eltype(x),sqrt(2*p[2]))]
end
## Gauss-Newton scheme
function GaussNewton!(xs, x, p; maxiter=8, backend=nothing)
for i=1:maxiter
x = step(x, p, backend)
@info i
@show x
push!(xs, x)
end
return xs, x
end
done(x,x2,p) = g(x2,p) < g(x,p)
function step(x, p, backend::Nothing, α=1//1)
x2 = deepcopy(x)
while !done(x,x2,p)
J = Jacobian(x,p)
d = -inv(J'*J)*J'*f(x,p)
copyto!(x2,x + α*d)
α = α//2
end
return x2
end
When we run the algorithm, we find the global minimum after about the 7th iteration.
xs = [x₀]
GaussNewton!(xs, x₀, p)
# output:
[ Info: 1 ]
x = [0.125, -0.08750000000000001]
[ Info: 2 ]
x = [0.234375, -0.047265625000000006]
[ Info: 3 ]
x = [0.4257812499999995, 0.06800537109374968]
[ Info: 4 ]
x = [0.5693359374999986, 0.21857223510742047]
[ Info: 5 ]
x = [0.784667968749996, 0.5165503501892037]
[ Info: 6 ]
x = [0.9999999999999961, 0.9536321163177449]
[ Info: 7 ]
x = [0.9999999999999989, 0.9999999999999999]
[ Info: 8 ]
x = [1.0, 1.0]
If computing the Jacobian by hand is too cumbersome (or not possible for other reasons), we can compute it using finite differences. Within the AbstractDifferentiation API, we can directly define, for instance, the Jacobian of FiniteDifferences.jl as a new primitive operation.
## FiniteDifferences
struct FDMBackend{A} <: AD.AbstractFiniteDifference
alg::A
end
FDMBackend() = FDMBackend(central_fdm(5, 1))
const fdm_backend = FDMBackend()
# Minimal interface
AD.@primitive function jacobian(ab::FDMBackend, f, xs...)
return FiniteDifferences.jacobian(ab.alg, f, xs...)
end
# AD Jacobian returns tuple
# df_dx = AD.jacobian(fdm_backend, f(x,p), x₀, p)[1]
# df_dp = AD.jacobian(fdm_backend, f(x,p), x₀, p)[2]
@test AD.jacobian(fdm_backend, x->f(x,p), x₀)[1] ≈ Jacobian(x₀, p)
@test AD.jacobian(fdm_backend, f, x₀, p)[1] ≈ Jacobian(x₀, p)
After overloading the step function, we can run the Gauss-Newton algorithm as follows:
function step(x, p, backend, α=1//1)
x2 = deepcopy(x)
while !done(x,x2,p)
J = AD.jacobian(backend, f, x, p)[1]
d = -inv(J'*J)*J'*f(x,p)
copyto!(x2,x + α*d)
α = α//2
end
return x2
end
xs = [x₀]
GaussNewton!(xs, x₀, p, backend=fdm_backend)
If we want to use reverse-mode AD instead, for example via Zygote.jl, a natural choice for the primitive is to define the pullback function. AbstractDifferentiation then generates the associated code to compute the Jacobian for us.
## Zygote
struct ZygoteBackend <: AD.AbstractReverseMode end
const zygote_backend = ZygoteBackend()
AD.@primitive function pullback_function(ab::ZygoteBackend, f, xs...)
return function (vs)
# Supports only single output
_, back = Zygote.pullback(f, xs...)
if vs isa AbstractVector
back(vs)
else
@assert length(vs) == 1
back(vs[1])
end
end
end
##
@test minimum(AD.jacobian(fdm_backend, f, x₀, p) .≈ AD.jacobian(zygote_backend, f, x₀, p))
xs = [x₀]
GaussNewton!(xs, x₀, p, backend=zygote_backend)
Typically, reverse-mode AD is only beneficial for functions $f:\mathbb{R}^N\rightarrow\mathbb{R}^M$ where $M \ll N$, thus it is also a good idea to compare the performance with respect to forward-mode AD (ForwardDiff.jl)
## ForwardDiff
struct ForwardDiffBackend <: AD.AbstractForwardMode end
const forwarddiff_backend = ForwardDiffBackend()
AD.@primitive function pushforward_function(ab::ForwardDiffBackend, f, xs...)
# jvp = f'(x)*v, i.e., differentiate f(x + h*v) wrt h at 0
return function (vs)
if xs isa Tuple
@assert length(xs) <= 2
if length(xs) == 1
(ForwardDiff.derivative(h->f(xs[1]+h*vs[1]),0),)
else
ForwardDiff.derivative(h->f(xs[1]+h*vs[1], xs[2]+h*vs[2]),0)
end
else
ForwardDiff.derivative(h->f(xs+h*vs),0)
end
end
end
##
@test minimum(AD.jacobian(fdm_backend, f, x₀, p) .≈ AD.jacobian(forwarddiff_backend, f, x₀, p))
xs = [x₀]
GaussNewton!(xs, x₀, p, backend=forwarddiff_backend)
where we have used that the Jacobian-vector product $f’(x)v$, i.e., the primitives of forward-mode AD, can be computed by differentiating $f(x + hv)$ with respect to $h$ at 0.
Many AD packages, such as Zygote, have troubles with mutating functions. Enzyme.jl is one of the exceptions. Additionally, it is very fast and has further improved the performance of the adjoints implemented within the DiffEqSensitivity package.
## Enzyme
struct EnzymeBackend{T1,T2,T3,T4} <: AD.AbstractReverseMode
out::T1
λ::T2
∂f_∂x::T3
∂f_∂p::T4
end
out = zero(x₀)
λ = zero(x₀)
∂f_∂x = zero(x₀)
∂f_∂p = zero(p)
const enzyme_backend = EnzymeBackend(out,λ,∂f_∂x,∂f_∂p)
AD.@primitive function pullback_function(ab::EnzymeBackend, f, xs...)
return function (vs)
# enzyme works only with inplace functions
if !(vs isa AbstractVector)
@assert length(vs) == 1 # Supports only single output
vs = vs[1]
end
if xs isa Tuple
@assert length(xs) == 2 # hard-coded for use case with two inputs
x₀ = xs[1]
p = xs[2]
end
@unpack out, λ, ∂f_∂x, ∂f_∂p = ab # cached in the struct, could also be created in here
∂f_∂x .*= false
∂f_∂p .*= false
out .*= false
copyto!(λ, vs)
autodiff(Duplicated(out, λ), Duplicated(x₀, ∂f_∂x), Duplicated(p, ∂f_∂p)) do _out,_x, _p
f(_out,_x,_p)
end
return (∂f_∂x,∂f_∂p)
end
end
AD.isinplace(ab::EnzymeBackend) = true
AD.primalvalue(ab::EnzymeBackend, nothing, f, xs) = (f(ab.out,xs...);return ab.out)
##
@test minimum(AD.jacobian(fdm_backend, f, x₀, p) .≈ AD.jacobian(enzyme_backend, f, x₀, p))
xs = [x₀]
GaussNewton!(xs, x₀, p, backend=enzyme_backend)
Note that we have declared the Enzyme backend as inplace (which is important for internal control flow) and specified a primalvalue function returning the primal value of the forward pass.
Some current glitches
First, the push forward of a tuple of vectors, e.g., $(v_1, v_2)$, for a function with several input arguments is currently ambiguous. While AD.jacobian primitives and AD.pullback_function primitives interpret the push forward of our $f$ function as
$$ \left(\frac{\partial f(x_0,p)}{\partial x} v_1 , \frac{\partial f(x_0,p)}{\partial p} v_2 \right), $$
AD.pushforward_function primitives compute
$$ \frac{\partial f(x_0,p)}{\partial x} v_1 + \frac{\partial f(x_0,p)}{\partial p} v_2. $$
# pushforward_function wrt to multiple vectors is currently ambiguous
vs = (randn(2), randn(2))
res1 = AD.pushforward_function(fdm_backend, f, x₀, p)(vs)
res2 = AD.pushforward_function(forwarddiff_backend, f, x₀, p)(vs)
@test res2 ≈ res1[1] + res1[2]
Thus, we currently solve this issue by augmenting the input in the case of AD.pushforward_function primitives.
res2a = AD.pushforward_function(forwarddiff_backend, f, x₀, p)((vs[1], zero(vs[2])))
res2b = AD.pushforward_function(forwarddiff_backend, f, x₀, p)((zero(vs[1]), vs[2]))
@test res2a ≈ res1[1]
@test res2b ≈ res1[2]
The plural “primitives” is used here because we may have different pushforward_function primitives for different backends. For instance, we can define an additional pushforward_function primitive for FiniteDifferences by:
struct FDMBackend2{A} <: AD.AbstractFiniteDifference
alg::A
end
FDMBackend2() = FDMBackend2(central_fdm(5, 1))
const fdm_backend2 = FDMBackend2()
AD.@primitive function pushforward_function(ab::FDMBackend2, f, xs...)
return function (vs)
FDM.jvp(ab.alg, f, tuple.(xs, vs)...)
end
end
Second, to avoid misunderstandings for the output of a Hessian of a function with several input arguments, we allow only single input arguments to the Hessian function.
# Hessian only defined with respect to single input variable
@test_throws AssertionError H1 = AD.hessian(forwarddiff_backend, g, x₀, p)
H1 = AD.hessian(forwarddiff_backend, x->g(x,p), x₀)
H2 = AD.hessian(forwarddiff_backend, p->g(x₀,p), p)
Third, computing the Hessian requires to nest AD/backend calls. This can lead to failure if one tries to use Zygote over Zygote. To solve this problem, we have implemented a HigherOrderBackend that takes a tuple containing multiple backends (because, for example, using ForwardDiff over Zygote is perfectly fine).
# Hessian might fail if AD system calls must not be nested (e.g. Zygote over Zygote)
backends = AD.HigherOrderBackend((forwarddiff_backend,zygote_backend))
H3 = AD.hessian(backends, x->g(x,p), x₀)
Outlook
There are many other use cases, e.g.,
- Sensitivity analysis of differential equations requires vector-Jacobian products for adjoint methods and Jacobian-vector products for tangent methods.
- The Newton–Raphson method for rootfinding requires the gradient in the case of scalar function $f:\mathbb{R}\rightarrow\mathbb{R}$ and the Jacobian in case of $N$ (nonlinear) equations, i.e., finding the zeros of $f:\mathbb{R}^N\rightarrow\mathbb{R}^N$.
- The Newton method in optimization requires the computation of the Hessian.
AbstractDifferentiation.jl is by no means complete yet. We are still in the very early stages, but we hope to make significant progress in the coming weeks. Some of the next steps are:
- fixing remaining bugs, e.g., with respect to the computation of the Hessian and
- adding AD/Finite Differentiation packages such as Diffractor.
If you have any questions or comments, please don’t hesitate to contact me!
-
Mike Innes, Alan Edelman, et al., arXiv preprint arXiv:1907.07587 (2019). ↩︎
-
Jeff Bezanson, Stefan Karpinski, et al., arXiv preprint arXiv:1209.5145 (2012). ↩︎
-
Chris Rackauckas, The Winnower 8, DOI: 10.15200/winn.156631.13064 (2019). ↩︎
-
Chris Rackauckas, Yingbo Ma, et al., arXiv preprint arXiv:2001.04385 (2020). ↩︎ ↩︎
-
Raj Dandekar, Chris Rackauckas, et al., Patterns 1, 100145 (2020). ↩︎
-
Frank Schäfer, Michal Kloc, et al., Mach. Learn.: Sci. Technol. 1, 035009 (2020). ↩︎
-
Frank Schäfer, Pavel Sekatski, et al., Mach. Learn.: Sci. Technol. 2, 035004 (2021). ↩︎